Downstream Assay–Driven Buffer Selection in Bead Mill Homogenization
January 30, 2026Efficient and reproducible tissue homogenization is a critical first step in protein analysis workflows, as incomplete lysis can lead to biased results and reduced analytical sensitivity. To address this, many laboratories routinely rely on chemical lysis buffers, such as RIPA lysis buffer, that contain detergents and chaotropic agents designed to disrupt cellular membranes and solubilize proteins. While effective, these reagents present notable limitations, as components such as SDS, Triton™ X-100, and deoxycholate are known to interfere with a broad range of downstream assays, from routine protein absorbance measurements to more complex enzyme-based reactions, immunoassays, and mass spectrometry–based analyses. 1,2,3.
As a result, researchers are often faced with a trade-off between achieving efficient lysis and maintaining compatibility with downstream applications. In many cases, samples processed in lysis buffers must undergo additional cleanup or buffer exchange steps, increasing workflow complexity, sample loss, and variability. This challenge has driven interest in alternative homogenization strategies that minimize reliance on chemical lysis while still ensuring complete cell disruption.
In addition to assay interference, chemical lysis alone may require extended incubation times and repeated mixing to achieve sufficient tissue disruption, particularly for complex or heterogeneous samples. Even with prolonged exposure to detergents, incomplete homogenization can occur, leaving intact tissue fragments or partially lysed cells. This lack of uniformity can introduce sample-to-sample variability and compromise reproducibility, especially when processing multiple samples in parallel.
Bead-based mechanical homogenization offers a powerful solution by physically disrupting tissues and cells through high-energy bead milling, independent of buffer composition. When sufficient mechanical energy is applied, cellular membranes can be efficiently ruptured in simple, assay-compatible buffers such as PBS.
In this application note, we evaluate the performance of the Omni Bead Ruptor Elite™ for homogenizing chicken tissue samples in PBS compared to a commonly used detergent-based lysis buffer (RIPA). Protein yield was assessed using a bicinchoninic acid (BCA) assay, while lysis efficiency was independently verified using acridine orange/ propidium iodide (AO/PI) staining to assess cellular integrity post-homogenization. By decoupling mechanical lysis from chemical solubilization, this study demonstrates how bead milling enables flexible sample preparation workflows, allowing researchers to select buffers based on downstream assay requirements rather than lysis efficiency alone.
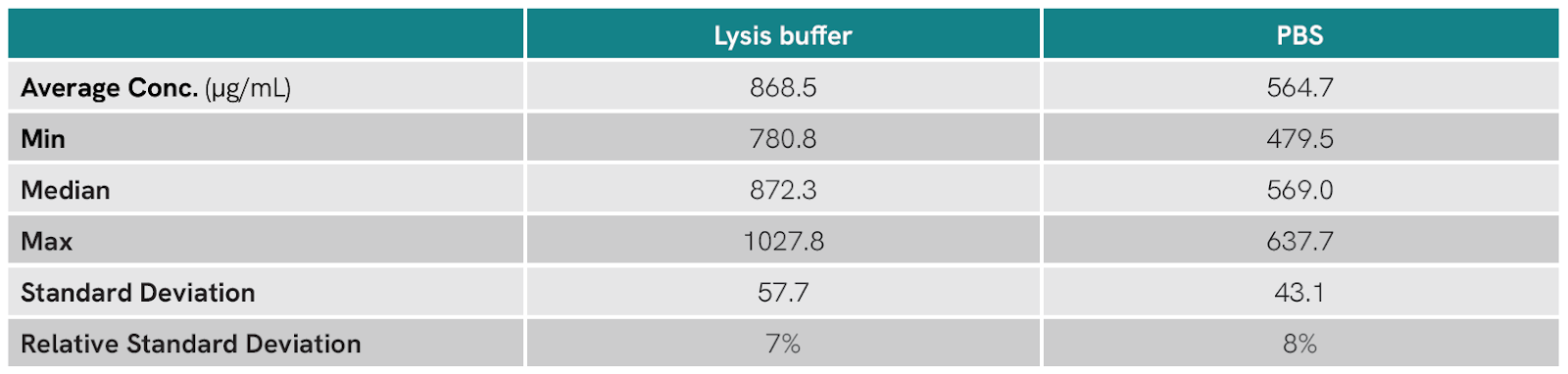
Table 1. Summary statistics of protein concentration measured by BCA assay for chicken tissue homogenized in RIPA lysis buffer or PBS using the Bead Ruptor Elite.
To View the Full Application Note Please Complete the Form Below
2 Wiśniewski, J. R. (2018). Filter-aided sample preparation for proteome analysis. In Microbial proteomics: methods and protocols (pp. 3-10). New York, NY: Springer New York. www.revvity.com Copyright ©2026, Revvity, Inc. All rights reserved. Revvity is a trademark of Revvity, Inc. All other trademarks are the property of their respective owners.
